Legrand Ebola SEIHFR#
Legrand_Ebola_SEIHFR()
A commonly used model in the literature to capture the dynamics of Ebola outbreaks is the SEIHFR model proposed by Legrand et al 7. There are two extra compartments on top of the SEIR: \(H\) for hospitializations and \(F\) for funerals. A total of ten parameters (with some describing the inverse) are required for the model.
Symbol |
Process |
|---|---|
\(\beta_{I}\) |
Transmission rate in community |
\(\beta_{H}\) |
Transmission rate in hospital |
\(\beta_{F}\) |
Transmission rate in funeral |
\(\gamma_{I}\) |
(inverse) Onset to end of infectious |
\(\gamma_{D}\) |
(inverse) Onset to death |
\(\gamma_{H}\) |
(inverse) Onset of hospitilization |
\(\gamma_{F}\) |
(inverse) Death to burial |
\(\alpha\) |
(inverse) Duration of the incubation period |
\(\theta\) |
Proportional of cases hospitalized |
\(\delta\) |
Case–ftality ratio |
The (inverse) denotes the parameter should be inverted to make
epidemiological sense. We use the parameters in their more natural from
in Legrand_Ebola_SEIHFR() and replace all the \(\gamma\)’s with
\(\omega\)’s, i.e. \(\omega_{i} = \gamma_{i}^{-1}\) for \(i \in \{I,D,H,F\}\).
We also used \(\alpha^{-1}\) in our model instead of \(\alpha\) so that
reading the parameters directly gives a more intuitive meaning. There
are five additional parameters that is derived. The two derived case
fatality ratio as
with an adjusted hospitalization parameter
and the derived infectious period
Now we are ready to state the full set of ODEs,
with \(\beta_{F}(t) = \beta_{F}\) if \(t > c\) and \(0\) otherwise. We use a slightly modified version by replacing the delta function with a sigmoid function namely, the logistic function
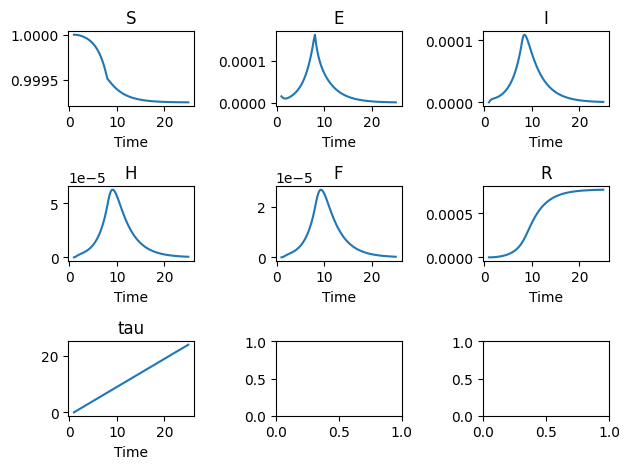
A brief example is given here:
import numpy
from pygom import common_models
x0 = [1.0, 3.0/200000.0, 0.0, 0.0, 0.0, 0.0, 0.0]
t = numpy.linspace(1, 25, 100)
ode = common_models.Legrand_Ebola_SEIHFR([('beta_I',0.588),
('beta_H',0.794),
('beta_F',7.653),
('omega_I',10.0/7.0),
('omega_D',9.6/7.0),
('omega_H',5.0/7.0),
('omega_F',2.0/7.0),
('alphaInv',7.0/7.0),
('delta',0.81),
('theta',0.80),
('kappa',300.0),
('interventionTime',7.0)])
ode.initial_values = (x0, t[0])
solution = ode.integrate(t)
ode.plot()
Note
We have standardized the states so that the number of susceptible is 1 and equal to the whole population, i.e. \(N\) does not exist in our set of ODEs.