SIS#
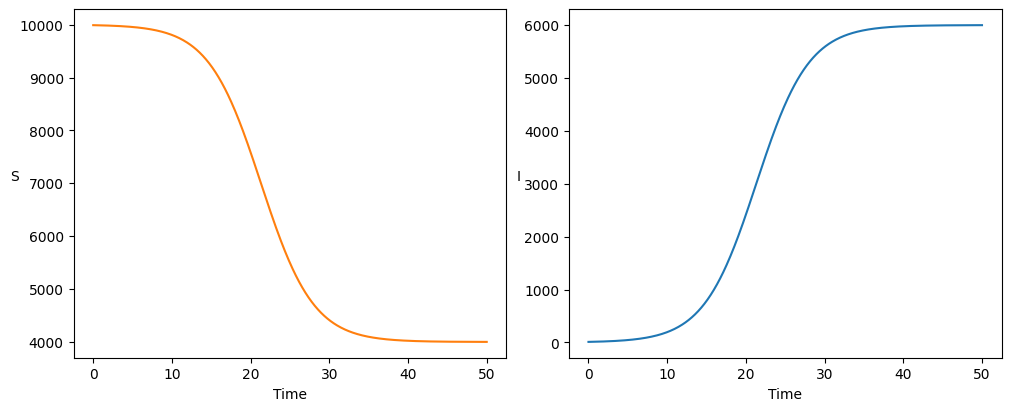
Perhaps the simplest epidemic model is a Susceptible-Infected-Susceptible (SIS) system, in which susceptible individuals may be infected and then do not have any immunity upon recovery.
\[\begin{split}\begin{aligned}
\frac{\mathrm{d}S}{\mathrm{d}t} &= -\frac{\beta S I}{N} + \gamma I \\
\frac{\mathrm{d}I}{\mathrm{d}t} &= \frac{\beta S I}{N} - \gamma I.
\end{aligned}\end{split}\]
We see how this evolves deterministically:
from pygom import common_models
import matplotlib.pyplot as plt
import numpy as np
import math
# Set up PyGOM object
n_pop=1e4
ode = common_models.SIS({'beta':0.5, 'gamma':0.2, 'N':n_pop})
# Initial conditions
i0=10
x0 = [n_pop-i0, i0]
# Time range and increments
tmax=50 # maximum time over which to run solver
dt=0.1 # timestep
n_timestep=math.ceil(tmax/dt) # number of iterations
t = np.linspace(0, tmax, n_timestep) # times at which solution will be evaluated
ode.initial_values = (x0, t[0])
solution=ode.solve_determ(t[1::])
After sufficiently long time, the system reaches an equilibrium state:
Show code cell source
f, axarr = plt.subplots(1,2, layout='constrained', figsize=(10, 4))
# Plot colours
colours=["C1", "C0"]
stateList=["S", "I"]
for i in range(0, 2):
axarr[i].plot(t, solution[:,i], color=colours[i])
axarr[i].set_ylabel(stateList[i], rotation=0)
axarr[i].set_xlabel('Time')
plt.show()